The Kaye laboratory focuses on the intersection of virology and cancer. About 20% of human cancers are caused by viruses. The lab is interested in understanding how virus infection can lead to cancer and developing strategies to disrupt this process. The lab has focused on a particular cancer virus, Kaposi’s sarcoma herpesvirus (KSHV, also known as human herpesvirus 8 (HHV8)). KSHV has a causative role in the malignancies Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL). KSHV also is highly associated with multicentric Castleman’s disease (MCD), an aggressive, lymphoma-like illness.
Similar to all herpesviruses, KSHV infection is lifelong. In most individuals, the virus causes no harm, and persists in harmony with its human host, where it is in equilibrium with the immune system. However, in some individuals, especially, when the immune system becomes compromised, such as in those with AIDS, virus infection can cause abnormal cell growth, resulting in malignancy (KS, PEL, or MCD).
To persist in its human host, KSHV establishes a latent infection in most cells it infects. During latency, the virus persists as a multicopy, circular, extrachromosomal, episome (plasmid). Although the virus encodes ~100 genes, only several are expressed during latent infection.
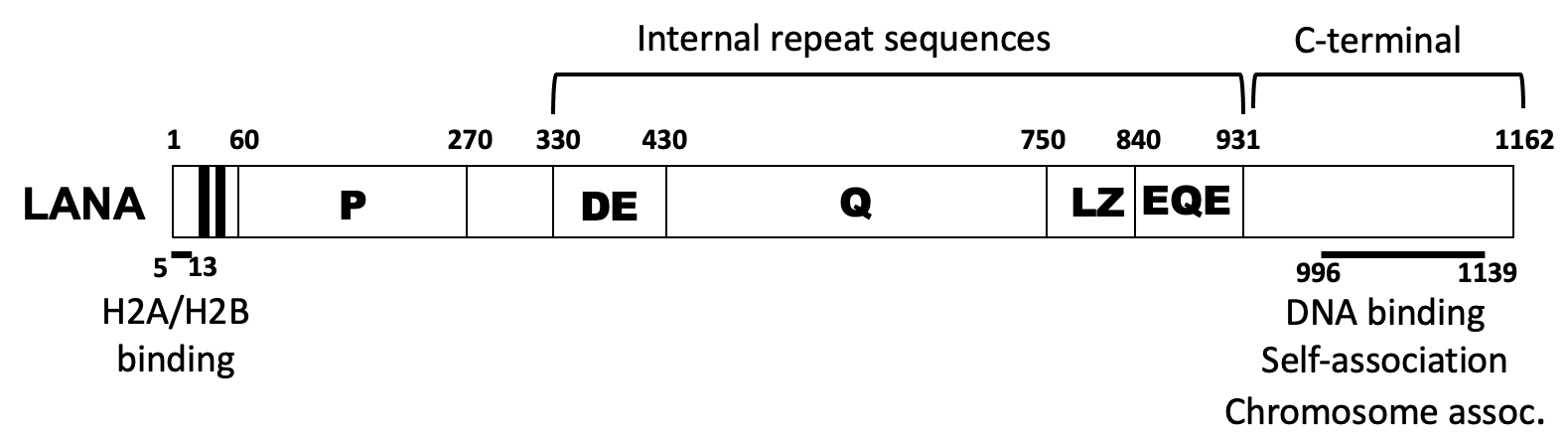
Schematic diagram of KSHV LANA. The proline-rich region (P), and internal repeat regions are indicated.
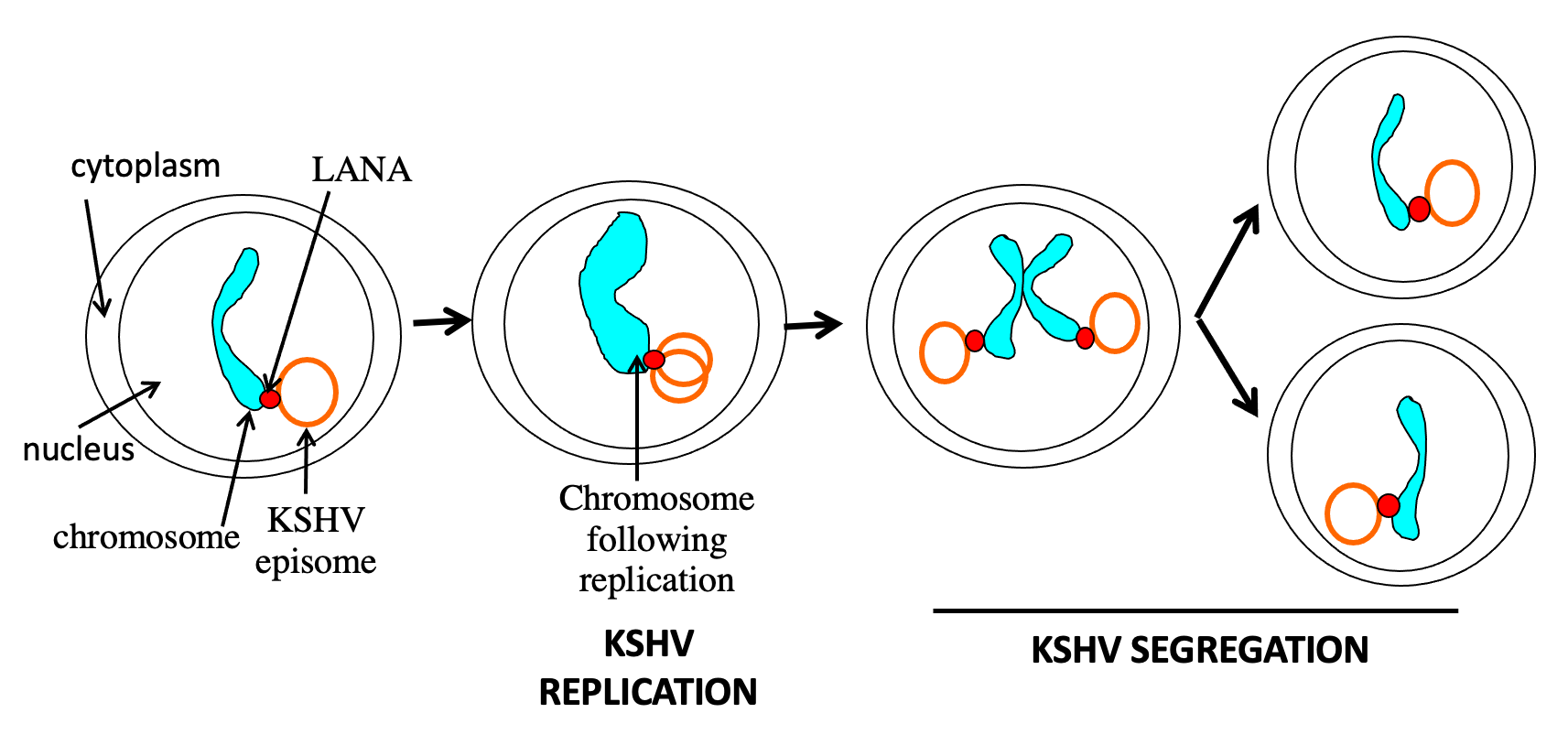
LANA mediated KSHV genome persistence in proliferating cells. LANA tethers the circular, viral genome (episome) to cell chromosomes. In S phase, the viral DNA replicates, along with host DNA. LANA enables replicated genomes to “hitchhike” along on mitotic chromosomes to ensure the viral DNA segregates to daughter cell nuclei.
The latency-associated nuclear antigen (LANA) is the predominant viral gene product expressed during latent infection. We discovered that LANA is responsible for viral episome maintenance. As a result, LANA is essential for viral persistence in latently infected, proliferating, cells. To permit episome maintenance, LANA mediates viral DNA replication and then segregates the replicated viral episomes to daughter cell nuclei by tethering the genomes to mitotic chromosomes. Thus, LANA serves as a molecular tether by simultaneously binding viral DNA and mitotic chromosomes. N-terminal LANA is the dominant chromosome attachment region, and we have found that C-terminal LANA also has a role in this process.
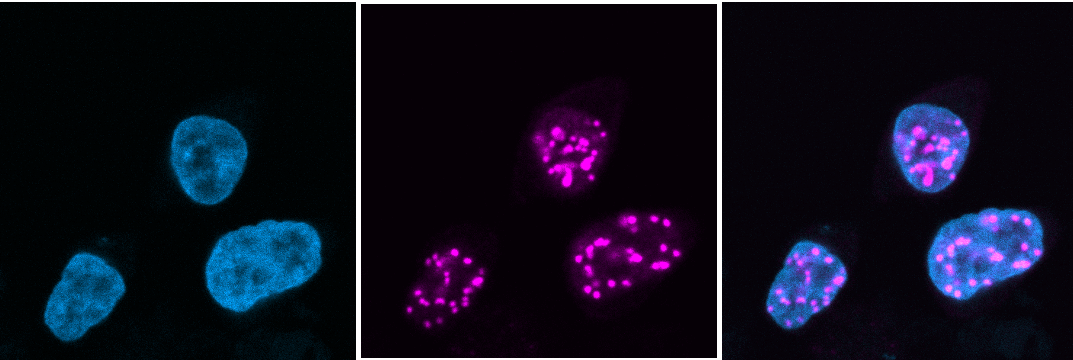
LANA (magenta) concentrates to dots at sites of KSHV genomes in nuclei of infected cells. Merge of DAPI (blue) staining (left) and LANA (center) is shown at right.
KSHV tumors are dependent on viral infection for viability. Since LANA is essential for viral persistence, strategies that disrupt LANA function would provide novel therapies for KSHV malignancy. Currently, there are no specific therapies for KSHV tumors, which often have poor prognoses.
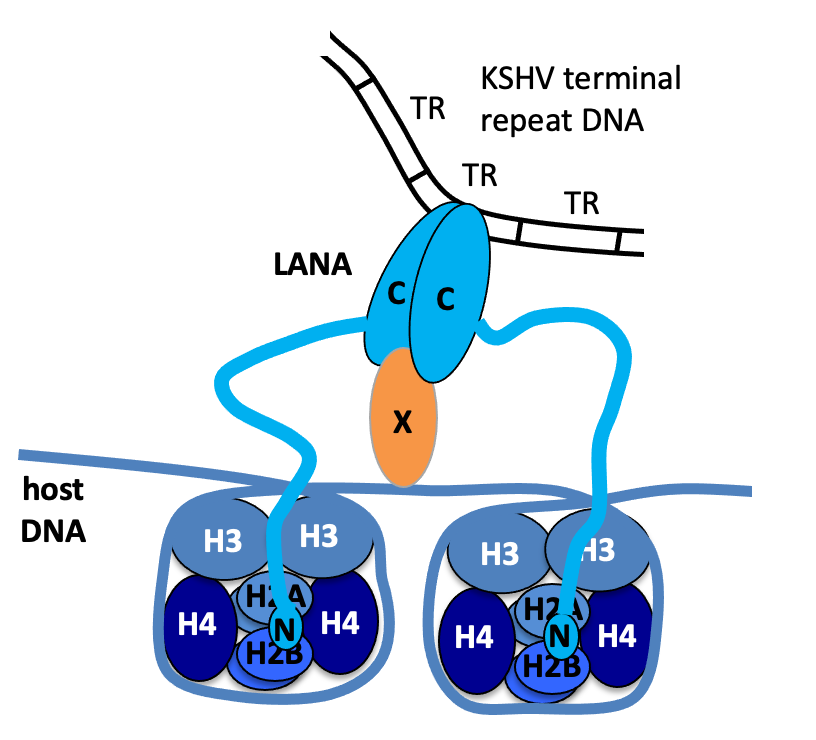
Model of LANA tethering. Amino-terminal LANA (N) binds to histones H2A/H2B. Carboxy-terminal LANA (C) forms a dimer, binds to KSHV terminal repeat DNA, and also binds to an as yet unindentified protein (X) that binds the chromosome.
We investigated how LANA attaches to viral DNA, and discovered that LANA binds specific sequence in viral terminal repeat DNA (TR) elements and showed that LANA was necessary and sufficient to act on TR DNA to mediate episome maintenance in the absence of other viral genes. LANA binds TR DNA through its carboxy-terminal domain, and LANA concentrates at KSHV episomal DNA due to LANA’s many binding sites present in the repetitive TR elements. LANA lacks enzymatic function and recruits host cell proteins to replicate DNA. As an example, we showed the LANA recruits the host cell DNA polymerase clamp (PCNA) loader, RFC, to mediate efficient replication.
LANA is an 1162 amino acid protein and its N- and C -terminal regions are essential for episome maintenance due to their roles in attachment to mitotic chromosomes and viral DNA, respectively. However, we have shown that deletion of the intervening ~1000 amino acid sequence induces severe deficiencies in episome maintenance. We have identified independent regions within this sequence that exert key functional effects. We recently discovered that the central, acidic repeat elements (DE) are highly homologous to the SET oncoprotein acidic domain reader sequence which preferentially binds unacetylated proteins, including p53. This sequence is necessary for efficient virus persistence, suggesting interaction with an unacetylated protein(s) partner might exert this effect.
LANA is a multifunctional protein, and also exerts transcriptional effects, which can be either enhancing or inhibitory, in different contexts. Accordingly, LANA interacts with multiple chromatin modifiers and histone binding proteins. We recently discovered that LANA recruits the mixed lineage leukemia 1 (MLL1) histone methyltransferase to viral DNA where it establishes H3K4me3 modifications on the TR DNA. We found that LANA interacts with MLL1 complex members, including WDR5, and regulates MLL1 activity. In addition, we found that MLL1 is important for KSHV latency establishment.
We discovered that LANA attaches to chromosomes by binding histones H2A/H2B and collaborated with Karolin Luger’s group to solve the x-ray crystal structure of N-terminal LANA complexed to the nucleosome. The structure demonstrates that N-LANA binds the conserved acidic patch at the interface of histones H2A/H2B on the nucleosome surface. Of interest, the acidic patch has subsequently been identified as a docking site for many host cell proteins.
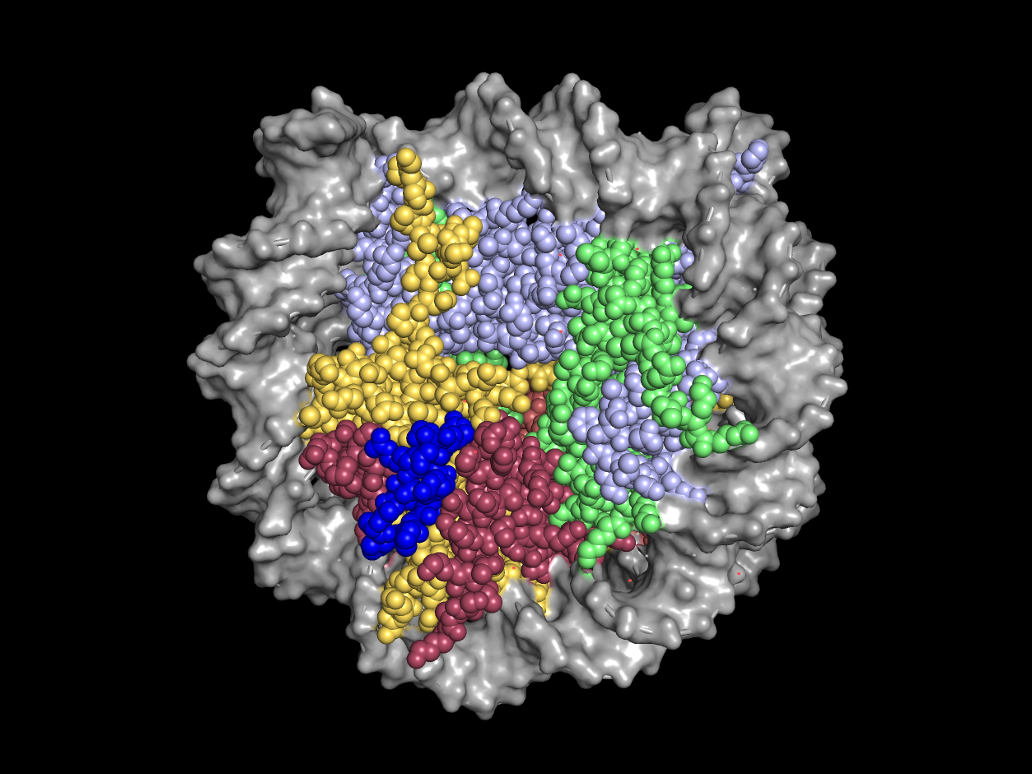
X-ray crystal structure of amino terminal LANA (blue) complexed with the nucleosome. A space-filling representation is shown. (PDB 1zla)
We found that carboxy-terminal LANA is the DNA binding domain and collaborated with Colin McVey and Maria Carrondo to solve its x-ray crystal structure. Notably, despite a lack of sequence homology, the structure shows high structural homology to the DNA binding domains of the Epstein-Barr virus EBNA1 and papillomavirus E2 proteins, which mediate episome maintenance for their respective viruses. The LANA DNA binding domain contains a basic patch on the surface opposite of the DNA binding interface, and we showed that this positive patch is important for efficient episome maintenance and interaction with BET family bromodomain proteins, including BRD4, although deficiencies in BET protein binding did not correlate with episome maintenance deficiency, suggesting independent functions.
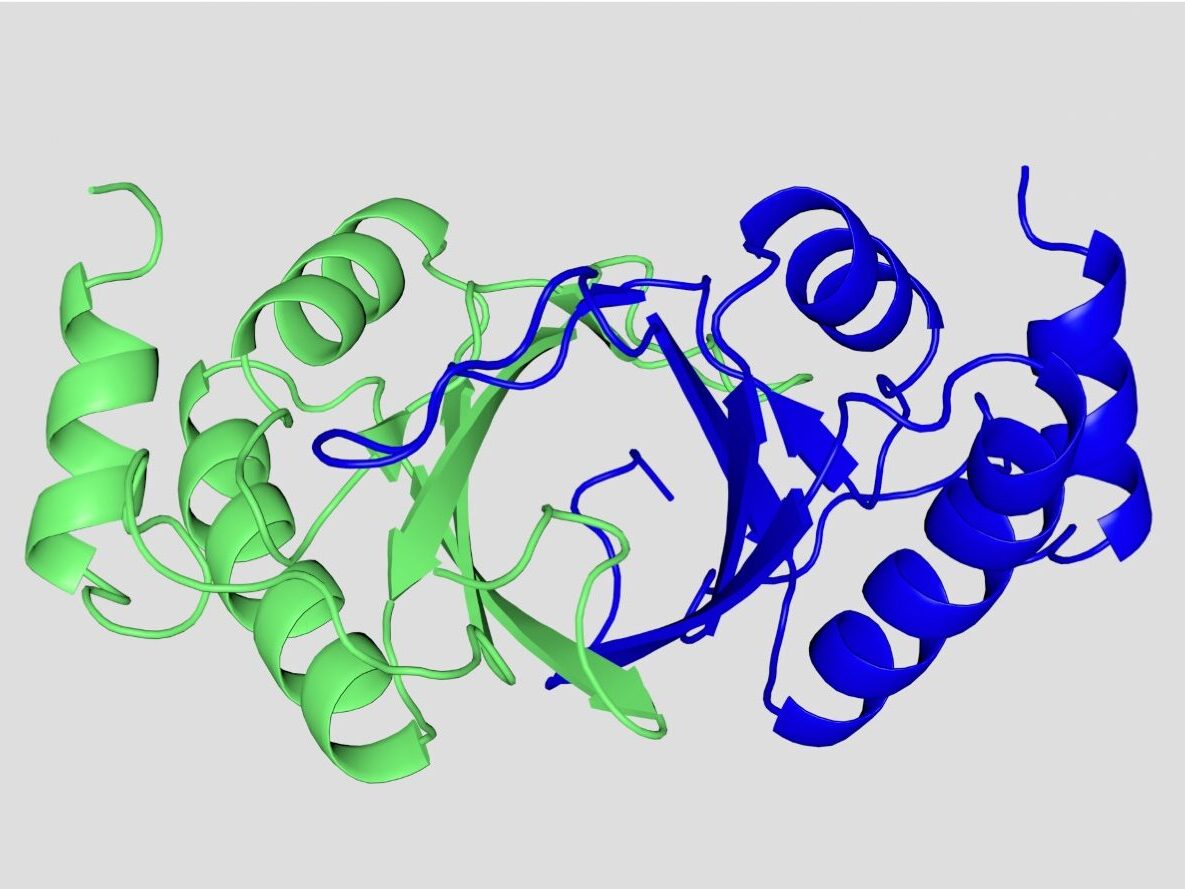
X-ray crystal structure of the dimeric C-terminal LANA DNA binding domain. Monomers are colored in blue and green. The ventral face is the DNA binding surface. (PDB 4blg)
We discovered that N-terminal LANA (downstream of the histone binding region) contains a WDR5 interacting motif (WIN), and collaborated with Colin McVey to solve the X-ray crystal structure of N-LANA complexed with WDR5, which reveals a potential MLL1 regulatory mechanism. N-terminal LANA inserts into the central WDR5 binding pocket, similar to the interaction of MLL1WIN with WDR5, suggesting competition for the site.
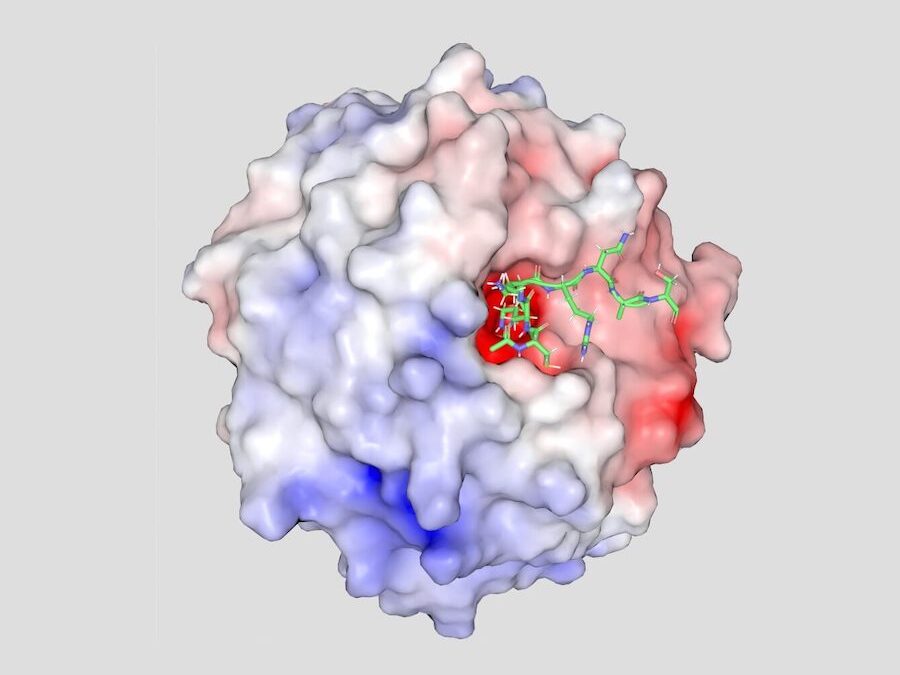
X-ray crystal structure of amino terminal LANA (green) complexed with WDR5. An electrostatic surface representation of WDR5 is shown. (PDB 7bcy)
KSHV’s natural host is humans. However, murine gammaherpesvirus 68 (MHV68) is a related virus that has a LANA homolog, and for which there is a well-characterized model for infection. Working with Pedro Simas’s group, we have shown that KSHV LANA can substitute for MHV68 LANA to allow establishment of viral latent infection in vivo. Use of this chimeric virus now allows us to investigate LANA function in vivo.
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The Nucleosomal Surface as a Docking Station for Kaposi’s Sarcoma Herpesvirus LANA. Science 2006;311:856-61.
- Correia B, Cerqueira SA, Beauchemin C, de Miranda MP, Li S, Ponnusamy R, Rodrigues L, Schneider TR, Carrondo MA*, Kaye KM*, Simas JP*, McVey CE*. Crystal Structure of the Gamma-2 Herpesvirus LANA DNA Binding Domain Identifies Charged Surface Residues Which Impact Viral Latency. Plos Pathogens 2013;9(10):e1003673.
- Li S, Tan M, Juillard J, Ponnusamy R, Correia B, Simas JP, Carrondo MA, McVey C, Kaye KM. The Kaposi’s Sarcoma Herpesvirus Latency-Associated Nuclear Antigen DNA Binding Domain Dorsal Positive Electrostatic Patch Facilitates DNA Replication and Episome Persistence. Journal of Biological Chemistry. 2015;290(47):28084-96.
- Habison, AC, Pires de Miranda M, Beauchemin C, Tan M, Cerqueira SA, Correia B, Ponnusamy R, Usherwood EJ, McVey CE, Simas JP, Kaye KM. Cross-Species Conservation of Episome Maintenance Provides a Basis for in Vivo Investigation of Kaposi’s Sarcoma Herpesvirus LANA. Plos Pathogens. 2017;13(9):e1006555.
- Juillard F, Pires de Miranda M, Li S, Franco A, Seixas AF, Liu B, Alvarez AL, Tan M, Szymula A, Kaye KM*, Simas JP*. KSHV LANA Acetylation-Selective Acidic Domain Reader Sequence Mediates Virus Persistence. Proc Natl Acad Sci U S A. 2020;117(36):22443-51.
- Tan M, Li S, Juillard F, Chitas R, Custodio TF, Xue H, Szymula A, Sun Q, Liu B, Alvarez AL, Chen S, Huang J, Simas JP, McVey CE, Kaye KM. MLL1 Is Regulated by KSHV LANA and Is Important for Virus Latency. Nucleic Acids Research. 2021;49(22):12895-911.